 |
|---|
 |
|---|
As constituent parts of image objects, superpixels can improve several higher-level operations. However, image segmentation methods might have their accuracy seriously compromised for reduced numbers of superpixels. We have investigated a solution based on the Iterative Spanning Forest (ISF) framework. In this letter, we present Dynamic ISF (DISF) — a method based on the following steps. (a) It starts from an image graph and a seed set with considerably more pixels than the desired number of superpixels. (b) The seeds compete among themselves, and each seed conquers its most closely connected pixels, resulting in an image partition (spanning forest) with connected superpixels. In step (c), DISF assigns relevance values to seeds based on superpixel analysis and removes the most irrelevant ones. Steps (b) and (c) are repeated until the desired number of superpixels is reached. DISF has the chance to reconstruct relevant edges after each iteration, when compared to region merging algorithms. As compared to other seed-based superpixel methods, DISF is more likely to find relevant seeds. It also introduces dynamic arc-weight estimation in the ISF framework for more effective superpixel delineation, and we demonstrate all results on three datasets with distinct object properties.
 |
 |
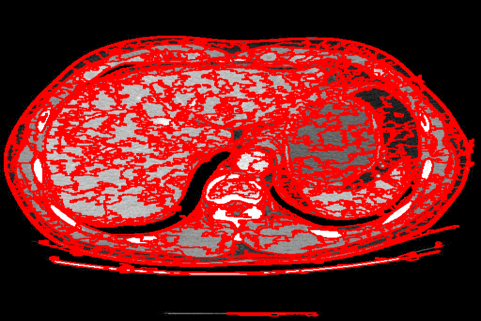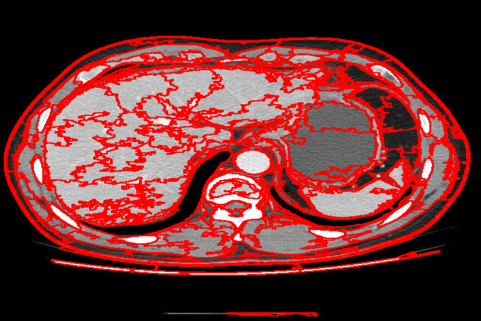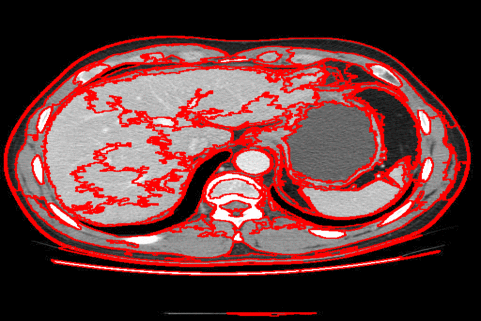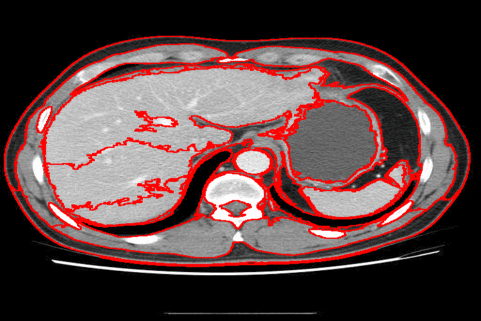 |
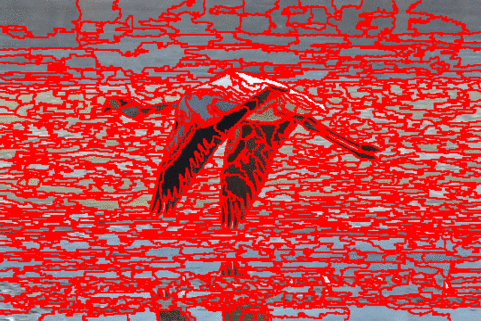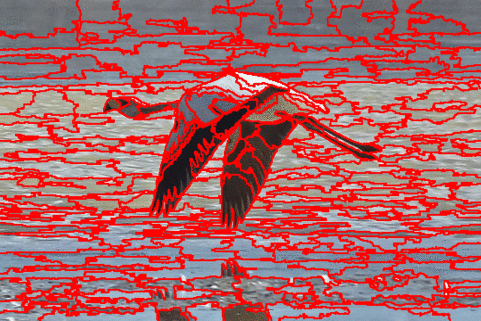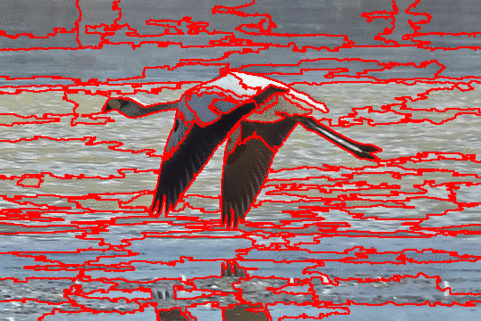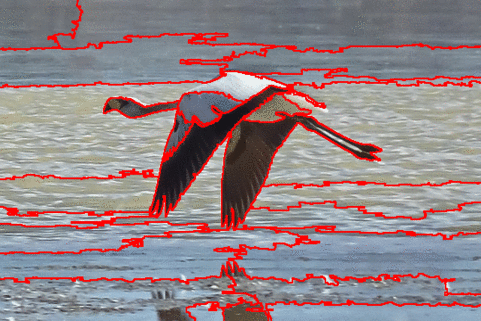
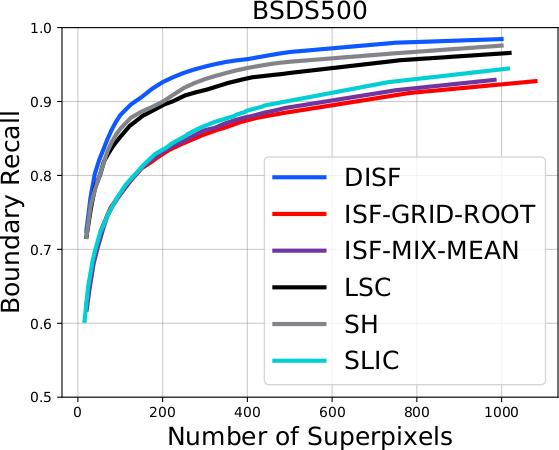
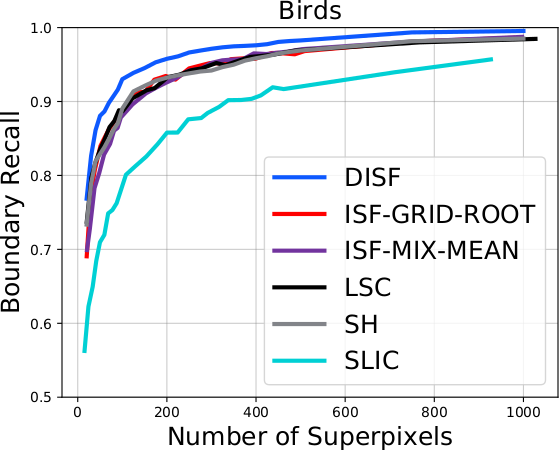
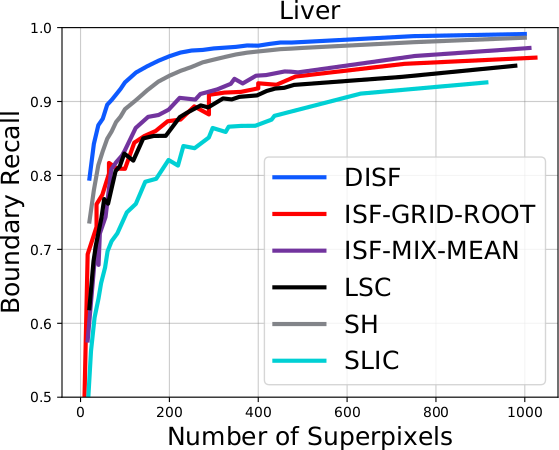
The DISF algorithm retains the most stable borders along its iterations until the number of desired superpixels achieved. Moreover, irrelevant superpixels (e.g., too small, or belongs to a homogeneous region) are identified and such that the object borders are minimally impacted. Such strategy permits a better boundary adherence and consistency in between different segmentation results, for distinct final number of superpixels (e.g.,1000, 500, 250, 100, 50 and 25), as the figures illustrate.
We evaluated our algorithm with different state-of-the-art methods:
| Linear Spectral Clustering (LSC) | Simple Linear Iterative Clustering (SLIC) |
|---|---|
| Li and Chen. “Superpixel segmentation using linear spectral clustering.” Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015 | Achanta et al. “SLIC superpixels compared to state-of-the-art superpixel methods.” IEEE Transactions on Pattern Analysis and Machine Intelligence, 2012 |
| Superpixel Hierarchy (SH) | Iterative Spanning Forest (ISF) |
|---|---|
| Wei et al. “Superpixel Hierarchy.” IEEE Transactions in Image Processing, 2018 | Vargas-Muñoz et al. “An iterative spanning forest framework for superpixel segmentation.” IEEE Transactions on Image Processing, 2019 |
Moreover, we evaluated the methods using the two most popular superpixel segmentation segmentation metrics:
| Boundary Recall (BR) | Under-Segmentation Error (UE) |
|---|---|
| Achanta et al. “SLIC superpixels compared to state-of-the-art superpixel methods.” IEEE Transactions on Pattern Analysis and Machine Intelligence, 2012. | Neubert and Protzel. “Superpixel benchmark and comparison.” Proceedings Forum Bildverarbeitung, 2012. |
And we considered three datasets from different domains:
| Birds | Liver | Berkeley Segmentation Dataset (BSDS500) |
|---|---|---|
| Mansilla and Miranda. “Oriented image foresting transform segmentation: Connectivity constraints with adjustable width.” Proceedings of the IEEE Conference on Graphics, Patterns and Images, 2016. | Vargas-Muñoz et al. “An iterative spanning forest framework for superpixel segmentation.” IEEE Transactions on Image Processing, 2019. | Arbelaez, et al. “Contour detection and hierarchical image segmentation.” IEEE Transactions on Pattern Analysis and Machine Intelligence, 2010 |
 |
 |
 |
|---|
As it can be seen in the charts, the DISF algorithm presents smoother curves due to its segmentation consistency. Moreover, its performance surpasses state-of-the-art’s in both terms of boundary recall and under-segmentation error. It is important to notice that the “starting point” of DISF is significantly better than the other methods, indicating better delineation for a small number of superpixels (which is often preferable).
This work was published in IEEE Signal Processing Letters journal and in the arXiv repository:
DISF was written in C, considering a Linux-based operational system, and its source code is avaliable for download (338KB). Moreover, it is possible to run the code in Python3, MATLAB, and Octave through wrappers (as illustrated by the demos). Although modifications can be performed for executing it in other environments, we CANNOT assure its proper compilation and execution in such. Please, read the README in the zipped file for details on how to compile and run our code. Finally, this implementation of DISF is licensed under Open Software License v3.0 (see LICENSE for more details).
We provide some example images from each dataset for download (1MB), for testing purposes.
Please, feel free to contact the authors for any unexpected behavior you might face (e.g., bugs). Moreover, if you’re interested in using our code, we appreciate if you cite this work in your project.